“Lab-on-Chip” Drug Discovery & Development
Microfluidic Devices and Artificial Cells
Many common problems of the drug discovery & development process can easily be addressed by microfluidic technology; it highly increases the potential of existing bioassays, chemical synthesis and separation technology. Our group is developing a novel on-chip approach, based on the integration of different micro-structured units, dedicated to the encapsulation of biological material, e.g. DNA and enzymes, into liposomes and the sorting of the resulting vesicles in a continuous stepwise manner according to their size and amount of material entrapped. These studies are aimed at developing new tools able to reduce the feature of a complex microbiological organism down to its basic properties such as compartmentalization and protein expression.
Scientists working on drug discovery & development are often searching for fast and robust methods to find, refine and test many potential drug candidates. A common trend, in order to reduce the costs and the time for the screening of nearly unlimited number of possibilities, relies on technological development of new tools for handling small liquid volumes, automation and high-throughput processing and analysis on a chip scale. Such small, enclosed laboratories can be useful to study chemical reactions in confined geometries (fig. 1). Moreover, miniaturization brings new scaling laws, e.g. enhanced surface per volume ratio, thus improving properties such as molecular diffusion or heat transport that can dramatically increase the efficiency and throughput potential.
We here want to emphasize particularly fluidic microchip systems, which provide the opportunity to perform reactions and analyses in a continuous manner. One of the promising possibilities of microfluidics is the compartmentalization of small liquid volumes within a carrier medium with high reproducibility. This microdroplet approach has been successfully used for chemical synthesis, cell analysis and cell-free protein expression [1-3]. It is important to underline that with classical methods for the generation of emulsions in batch (e.g. magnetic stir bar), the individual droplets cannot be accurately addressed and handled; therefore, automation and sorting are limited, whereas on a microfluidic platform many manipulation processes are available for fission, fusion and sorting of droplets. The microfluidic approach is compatible with high-throughput sequential detection and hence, allows easy individual droplet sorting and characterization [4].
Although the microdroplet concept is extremely valuable, wherever fast and homogeneous compartmentalization is required, it has some drawbacks, particularly concerning the water/carrier interface, which requires special care in order to prevent adsorption and loss of enclosed compounds. Therefore, we are interested in microchip-based creation of even more nature-like compartments, namely liposomes, which are - depending on their size and composition - reasonable model systems for cell organelles and cell.
The artificial cell strategies, based on the bottom-up approach, start with the construction of a minimal cell from the molecular level. One of the first challenges is to encapsulate genetic material and proteins into a vesicle composed of a phospholipid bilayer in non-denaturing conditions. Phospholipids are the main constituents of biological barriers; they offer many possibilities and great flexibility to functionalize and establish controllable exchanges between two phases [5]. Even though several methods for the generation of vesicles are available, a common problem for the batch productions is the generation of non-homogeneous vesicle populations often differing in size and amount of encapsulated material, which significantly reduces accuracy. Furthermore, to understand, biochemical properties of such artificial cells, methods to characterize individual liposomes, e.g. by means of flow cytometry, are required [6].
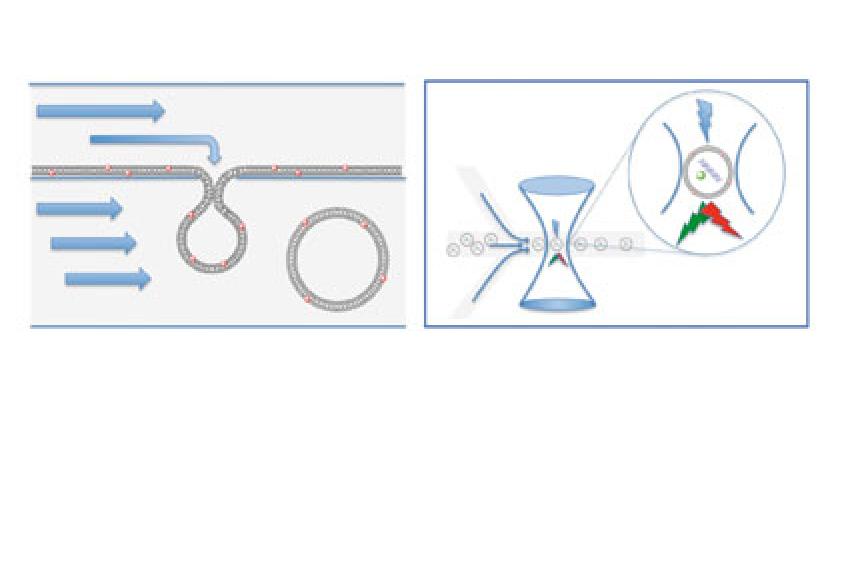
In our approach, we want to overcome the challenges by combining microfluidic-guided formation of liposomes with ultrasensitive fluorescence detection in a microsized flow cytometer. Recently, we have developed methods to form and tailor liposomes of different morphologies by means of an on-chip micro-extrusion process (fig. 2) [7]. Furthermore, we investigated the control on size and the influence of membrane composition on liposome formation, and we are now interested in fast, high-throughput determination of the properties, particularly entrapping capacity, of the liposomes.
For this purpose, a micro-cytometer chip, made of silicone elastomer and bonded to a glass slide, is used and mounted onto a home-build epifluorescent confocal setup (fig. 1), in which laser induced fluorescence of hydrodynamically focused individual vesicles was detected by aligning the centre of the micro channel, with the focused light. Fluorescence light emitted in the laser spot is collected through optical fibers and detected by avalanche photodiodes. Two different fluorescence colors - green and red - are collected simultaneously (fig. 2).
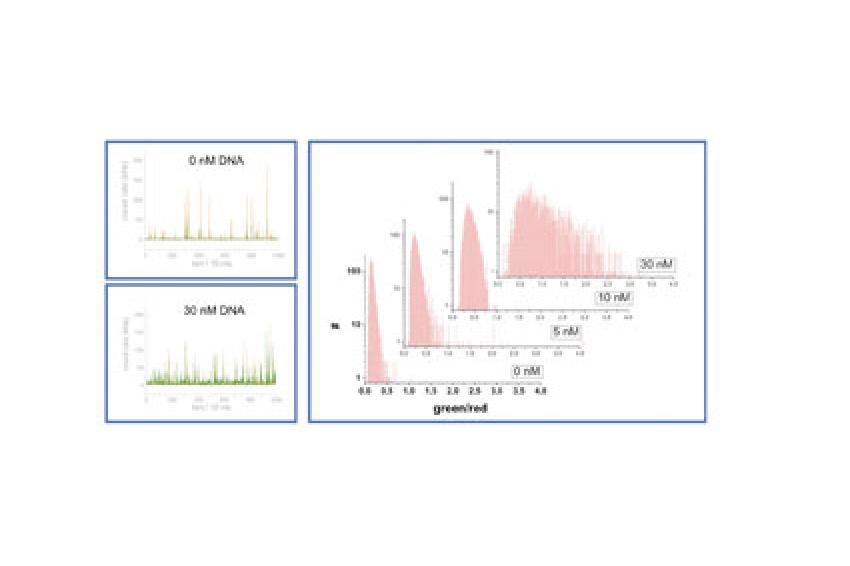
We produced several buffer suspensions of liposomes, labeling the membrane with the red fluorescent dye molecule (DiI-C18). The suspensions differed for the number (none, one, two, or more) of double strand DNA molecules labeled with rhodamine green (green fluorescent dye), which were entrapped in the liposome's lumens. Suspensions of liposomes and buffer solution for the sheath flow are introduced into the microchip by using syringe pumps (fig. 1).
The fluorescent red and green light generated upon excitation of the vesicles was acquired over time (with a throughput of several thousand of vesicles within a few minutes measuring time). Signals that are significantly above the background were analyzed, and the ratio of green to red fluorescence intensity was calculated. The ratio was compared among the different liposomes compositions containing none, one, two or more double strand DNA molecules. The results revealed a narrow distribution showing a significantly different threshold ratio for empty and filled liposomes. Liposomes entrapping a large number of DNA molecules revealed a very broad distribution. The resulting shift of the maxima of the threshold ratio distributions reflecting the increasing number of entrapped DNA molecule (fig. 3), clearly underlies the single molecule sensitivity of our method.
As a conclusion, microfluidic platform provide a perfect tool to form and analyze liposomes. Both modules, i.e. liposome formation by micro-extrusion and detection by micro-cytometry can be readily integrated on a single microdevice, which would allow for systematic studies on various parameters related to liposomes such as membrane properties, enclosure capacity and binding studies. Enclosing molecules in liposomes may have important applications for chemical reactions, biological screening, and evolutionary and single molecule studies. The exciting prospect of building an artificial cell that can perform specific tasks could be of great interest for pharmacology and medical diagnostics. Moreover, we believe that microchip liposome cytometry is a general approach for studying of membrane/membrane protein - drug interactions.
References
[1] Song H. et al.: Angew Chem Int Ed Engl 42, 768-772 (2003)
[2] Dittrich P. S. et al.: Chembiochem 6, 811-814 (2005)
[3] Link D.R. et al.: Angew Chem Int Ed Engl 45, 2556-2560 (2006)
[4] Fidalgo L.M. et al.: Angew Chem Int Ed Engl 48, 3665-3668 (2009)
[5] Noireaux V. and Libchaber A.: Proc Natl Acad Sci USA 101, 17669-17674 (2004)
[6] Sato K. et al.: J Biosci Bioeng 102, 171-178 (2006)
[7] Dittrich P.S. et al.: Lab Chip 6, 488-493 (2006)
Contact
ETH Zürich
Wolfgang-Pauli-Str. 10 -16
8093 Zürich
Switzerland
+41 44 632 1111
+41 44 632 1010